Launching the PROOF MVP
Creating and running bioinformatics workflows efficiently and consistently is crucial for research, however it can be tough to manage these workflows, especially when using different computing systems or platforms. To enable expanded use of workflows written using the WDL language, we have developed a new, open source tool, PROOF (Production Research On-ramp for Optimization and Feasibility). PROOF allows Fred Hutch staff to easily run their WDL workflows using our cluster, so you you can focus on your science, not the logistics of running and managing your computing tasks. It’s now easier than ever to get your workflows up and running and using a workflow language that is easily portable to other computing systems and platforms!
We’ve developed two easy-to-use ways of running your data processing WDL workflows on our local cluster.
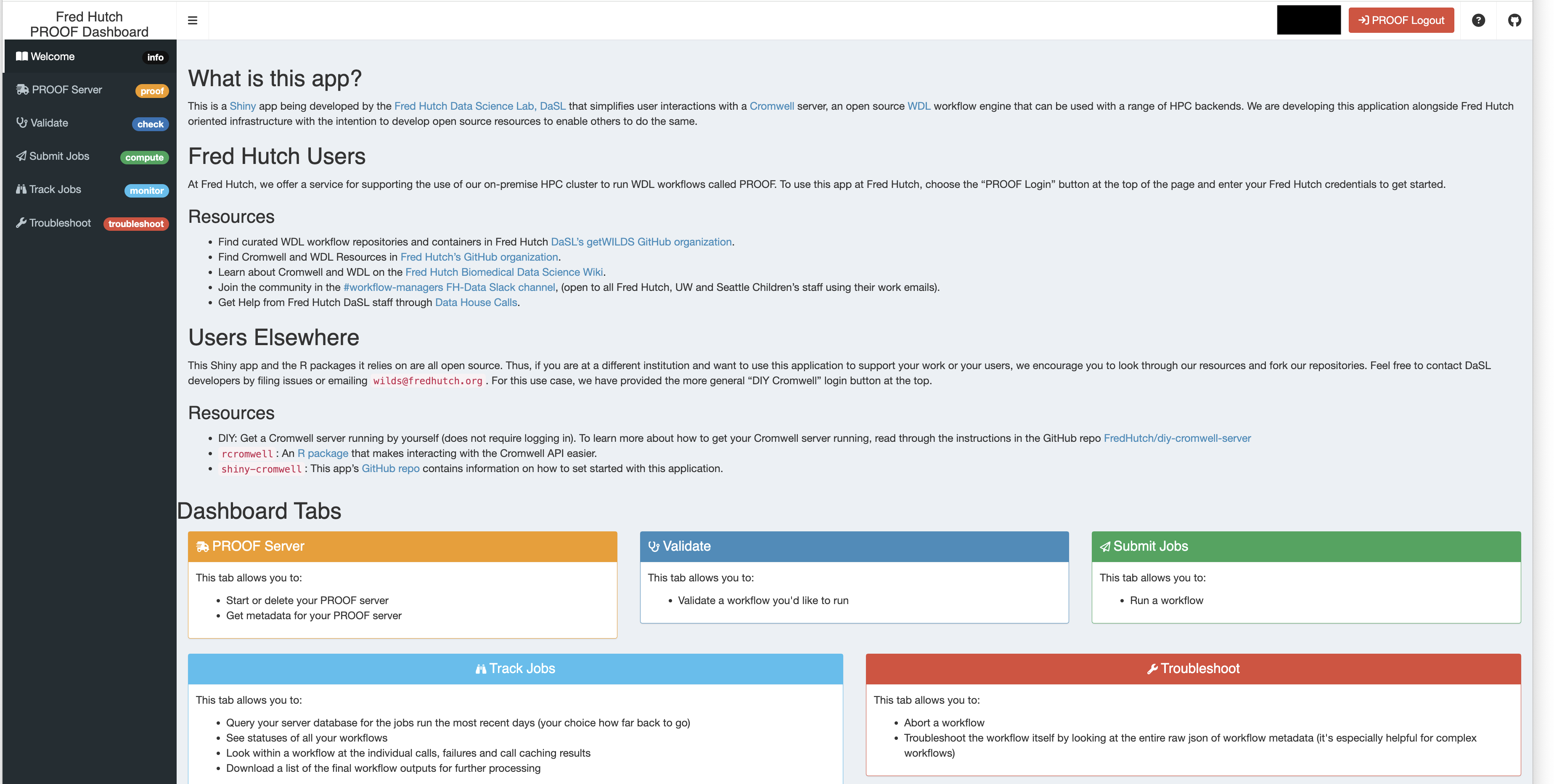
First, we have the PROOF Shiny application, an all-in-one application that allows you to validate, submit, and track WDL workflow jobs through an intuitive interface.
Second, for those who prefer to interact directly with their workflows through the command line, you can use PROOF via the packages proofr and rcromwell which allow you to validate, submit, and track your workflow jobs through the comfort of your R console.
Are you new to the workflow language or not sure where to get started? We have training materials and open workshops to help you get comfortable with writing WDLs, running workflows, and managing your work via PROOF.
These tools have all been released open source as resources for the wider community to leverage in their own data and computing ecosystem to support researchers more broadly than just Fred Hutch. Please fork our repositories, file issues or submit PRs if you’d like to collaborate or use these resources at your institution.