
Chapter 6 Iteration
Suppose that you want to repeat a chunk of code many times, but changing one variable’s value each time you do it. This could be modifying each element of a vector with the same operation, or analyzing a dataframe with different parameters.
There are three common strategies to go about this:
- Copy and paste the code chunk, and change that variable’s value. Repeat. This can be a starting point in your analysis, but will lead to errors easily.
- Use a
forloop to repeat the chunk of code, and let it loop over the changing variable’s value. This is popular for many programming languages, but the R programming culture encourages a functional way instead. - Functionals allow you to take a function that solves the problem for a single input and generalize it to handle any number of inputs. This is very popular in R programming culture.
6.1 For loops
A for loop repeats a chunk of code many times, once for each element of an input vector.
for (my_element in my_vector) {
chunk of code
}Most often, the “chunk of code” will make use of my_element.
6.1.0.1 We can loop through the indicies of a vector
The function seq_along() creates the indicies of a vector. It has almost the same properties as 1:length(my_vector), but avoids issues when the vector length is 0.
## [1] 1
## [1] 3
## [1] 5
## [1] 76.2 Functionals
A functional is a function that takes in a data structure and function as inputs and applies the function on the data structure, element by element. It maps your input data structure to an output data structure based on the function. It encourages the usage of modular functions in your code.

Or,
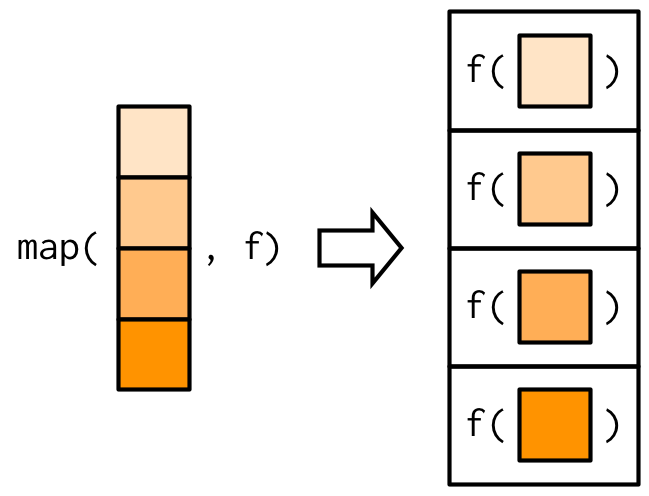
We will use the purrr package in tidyverse to use functionals. There is another set of functionals in Base-R called the apply family of functions that work very similarly. You can see the comparision of both tools here and here.
map() takes in a vector or a list, and then applies the function on each element of it. The output is always a list.
## [[1]]
## [1] 0
##
## [[2]]
## [1] 1.098612
##
## [[3]]
## [1] 1.609438
##
## [[4]]
## [1] 1.94591Lists are useful if what you are using it on requires a flexible data structure.
To be more specific about the output type, you can do this via the map_* function, where * specifies the output type: map_lgl(), map_chr(), and map_dbl() functions return vectors of logical values, strings, or numbers respectively.
For example, to make sure your output is a double (numeric):
## [1] 0.000000 1.098612 1.609438 1.945910All of these are toy examples that gets us familiar with the syntax, but we already have built-in functions to solve these problems, such as log(my_vector). Let’s see some real-life case studies.
6.3 Case studies
6.3.1 1. Loading in multiple files.
Suppose that we want to load in a few dataframes, and store them in a list of dataframes for analysis downstream.
We start with the filepaths we want to load in as dataframes.
The function we want to use to load the data in will be read_csv().
Let’s practice writing out one iteration:
6.3.1.1 To do this functionally, we think about:
What variable we need to loop through:
pathsThe repeated task as a function:
read_csv()The looping mechanism, and its output:
map()outputs lists.
6.3.2 2. Analyze a dataframe with different parameters.
Suppose you are working with the penguins dataframe:
## # A tibble: 6 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <fct> <fct> <dbl> <dbl> <int> <int>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## # ℹ 2 more variables: sex <fct>, year <int>and you want to look at the mean bill_length_mm for each of the three species (Adelie, Chinstrap, Gentoo).
Let’s practice writing out one iteration:
species_to_analyze = c("Adelie", "Chinstrap", "Gentoo")
penguins_subset = filter(penguins, species == species_to_analyze[1])
mean(penguins_subset$bill_length_mm, na.rm = TRUE)## [1] 38.791396.3.2.1 To do this functionally, we think about:
What variable we need to loop through:
c("Adelie", "Chinstrap", "Gentoo")The repeated task as a function: a custom function that takes in a specie of interest. The function filters the rows of
penguinsto the species of interest, and compute the mean ofbill_length_mm.The looping mechanism, and its output:
map_dbl()outputs (double) numeric vectors.
analysis = function(current_species) {
penguins_subset = dplyr::filter(penguins, species == current_species)
return(mean(penguins_subset$bill_length_mm, na.rm=TRUE))
}
map_dbl(c("Adelie", "Chinstrap", "Gentoo"), analysis)## [1] 38.79139 48.83382 47.504886.3.2.2 To do this with a for loop, we think about:
What variable we need to loop through:
c("Adelie", "Chinstrap", "Gentoo").Do we need to store the outcome of this loop in a data structure? Yes, a numeric vector.
At each iteration, what are we doing? Filter the rows of
penguinsto the species of interest, and compute the mean ofbill_length_mm.
outcome = rep(NA, length(species_to_analyze))
for(i in seq_along(species_to_analyze)) {
penguins_subset = filter(penguins, species == species_to_analyze[i])
outcome[i] = mean(penguins_subset$bill_length_mm, na.rm=TRUE)
}
outcome## [1] 38.79139 48.83382 47.504886.3.3 3. Calculate summary statistics on columns of a dataframe.
Suppose that you are interested in the numeric columns of the penguins dataframe.
penguins_numeric = penguins %>% select(bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g)and you are interested to look at the mean of each column. It is very helpful to interpret the dataframe penguins_numeric as a list, iterating through each column as an element of a list.
Let’s practice writing out one iteration:
## [1] 43.921936.3.3.1 To do this functionally, we think about:
What variable we need to loop through: the list
penguins_numericThe repeated task as a function:
mean()with the argumentna.rm = TRUE.The looping mechanism, and its output:
map_dbl()outputs (double) numeric vectors.
## bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## 43.92193 17.15117 200.91520 4201.75439Here, R is interpreting the dataframe penguins_numeric as a list, iterating through each column as an element of a list:
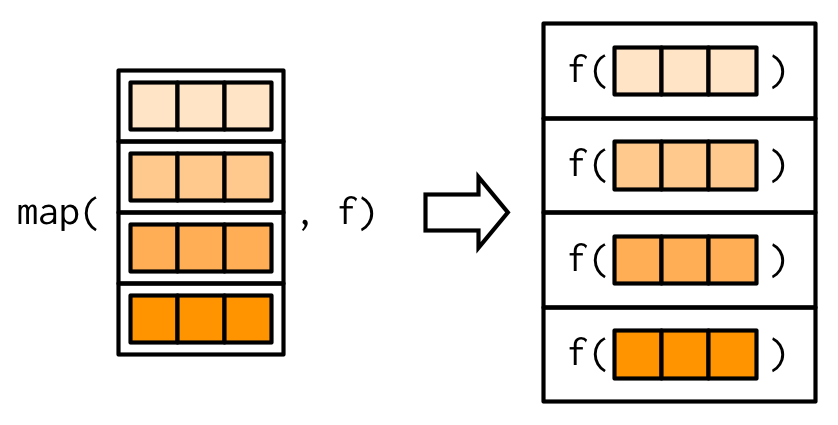
6.3.3.2 To do this with a for loop, we think about:
What variable we need to loop through: the elements of
penguins_numericas a list.Do we need to store the outcome of this loop in a data structure? Yes, a numeric vector.
At each iteration, what are we doing? Compute the mean of an element of
penguins_numeric.
result = rep(NA, ncol(penguins_numeric))
for(i in seq_along(penguins_numeric)) {
result[i] = mean(penguins_numeric[[i]], na.rm = TRUE)
}
result## [1] 43.92193 17.15117 200.91520 4201.754396.4 Exercises
You can find exercises and solutions on Posit Cloud, or on GitHub.
